Online Algorithm Tools to Enhance Precision Research
We provide a variety of leading classical algorithm tools, including protein structure prediction, molecular docking, virtual screening, PROTAC computation, MD simulation, reverse target screening, and more. Through a user-friendly web-based platform, you can easily operate without complex setup, quickly obtain detailed result analyses, and strongly support your research work.
Expert Customized Services to Meet Your R&D Needs
Primary Biotech brings together a team of seasoned experts from fields such as computer science, chemistry, biology, and medicine, with strong interdisciplinary backgrounds. We deeply understand the R&D needs in the life sciences field and are committed to providing personalized one-on-one customized services for every client. Based on your specific project requirements, we design tailored experimental plans to ensure optimal solutions for your research work.
Diverse Experimental Services to Support In-Depth Research
We offer comprehensive experimental services, including cell growth curve determination, cell invasion and migration assays, plate clone assays, stable cell line construction, cell cycle and apoptosis analysis, Real-Time PCR, Western Blot (WB), protein stability determination, Kd value determination, and more. Our professional team will customize experimental plans for you to ensure accurate and reliable results, providing strong data support for your research.
Comprehensive Software/Hardware System Setup Services for Efficient R&D
Primary Biotech provides complete computational biology software and hardware solutions, covering hardware facility configuration, environment setup, system management, database development, software installation, and debugging. Our professional team will provide customized solutions based on your needs to ensure the stable and efficient operation of your research platform and facilitate the smooth progress of your projects.
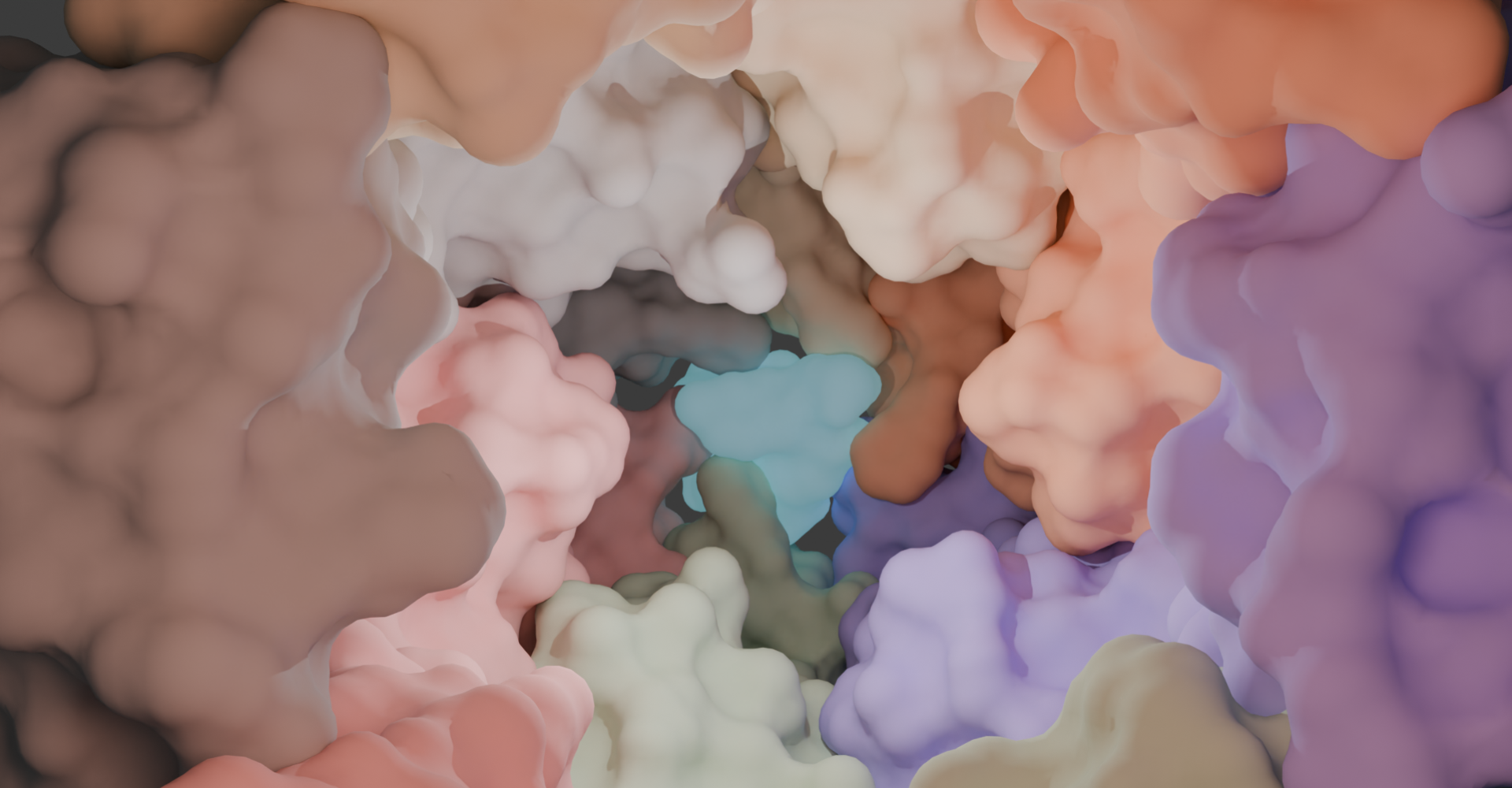
 Small Molecule Docking Tools
Small Molecule Docking Tools Macromolecule Docking Tools
Macromolecule Docking Tools
 Molecular Dynamics Simulation Tools
Molecular Dynamics Simulation Tools
 Common Small Tools
Common Small Tools